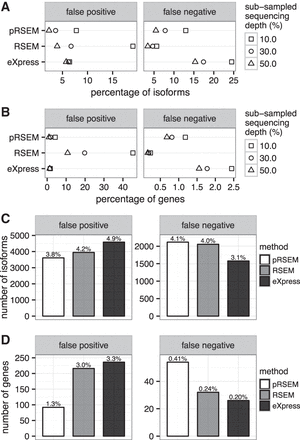
Data-driven simulations reveal that pRSEM has a lower false-positive rate (FPR) than RSEM and eXpress. (A,B) Shape plots (squares, circles, triangles) represent FPR and false negative rate (FNR) for isoforms (A) and genes (B) in subsampling experiments. (C,D) Bar plots show the number of isoforms (C) and genes (D) classified as false positives and false negatives from simulations at full sequencing depth. FPR and FNR are shown above each bar. A false positive is an isoform or gene with a true abundance <1 TPM and an estimated abundance ≥1 TPM. A false negative is an isoform or gene with a true abundance ≥1 TPM and an estimated abundance <1 TPM.











