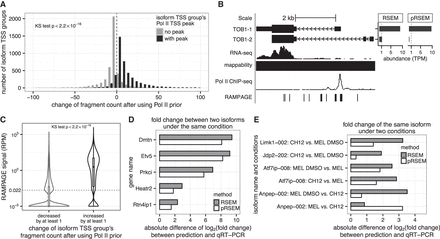
pRSEM more accurately allocates multimapping reads between isoforms. (A) Distributions of the change of fragment count between pRSEM and RSEM for isoform TSS groups with (black) and without (gray) Pol II TSS peaks. The two distributions are significantly different (P < 2.2 × 10−16, Kolmogorov-Smirnov test). (B) An example of multimapping read allocation. Shown are TOB1’s two protein-coding isoforms, mappability, RNA-seq signal, Pol II ChIP-seq signal, RAMPAGE signal, and estimated abundances from RSEM and pRSEM (top right). (C) Distributions of RAMPAGE signals for isoform TSS groups that have their fragment counts decreased by at least one (gray) or increased by at least one (black) after using a Pol II–informed prior. The dashed line denotes the signal corresponding to one RAMPAGE read. The two distributions are significantly different (P < 2.2 × 10−16, Kolmogorov-Smirnov test). A small fractional number (10−3) was added to the RAMPAGE signal for each isoform TSS group to allow display of zero signals on a log scale. Data shown in A through C are based on the K562 RNA-seq replicate one and RAMPAGE replicate one data sets. Isoform TSS groups in A and C are from genes that had “mixed” peak status and did not overlap or share reads with any other gene. Isoform TSS groups that were estimated to have abundance of less than one TPM according to both RSEM and pRSEM or that had fragment count changes between the two methods of less than one or larger than 100 were excluded. (D) Comparison of RSEM (gray) and pRSEM (white) estimated fold changes between a gene's two isoforms in the MEL cell line with fold changes measured by qRT-PCR. (E) Comparison of RSEM (gray) and pRSEM (white) estimated isoform fold changes between two conditions with fold changes measured by qRT-PCR.











