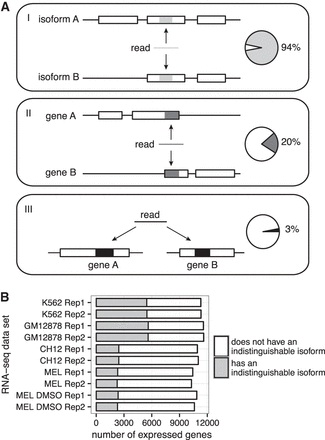
Multimapping reads are prevalent in human and mouse RNA-seq data. (A) Three classes of multimapping reads and the percentages of expressed genes (TPM ≥ 1) to which each class maps in the K562 replicate one data set. (B) Number of expressed genes that have indistinguishable isoforms. An indistinguishable isoform is one for which all potential RNA-seq fragments derived from it can align to other isoforms (for the calculations of distinguishability, see section I.B in the Supplemental Material). An expressed gene is required to have abundance of one TPM or more. Data are from the two RNA-seq replicates (“Rep1” and “Rep2”) of each of the human cell lines K562 and GM12878 and the mouse cell lines CH12, MEL, and MEL DMSO.











