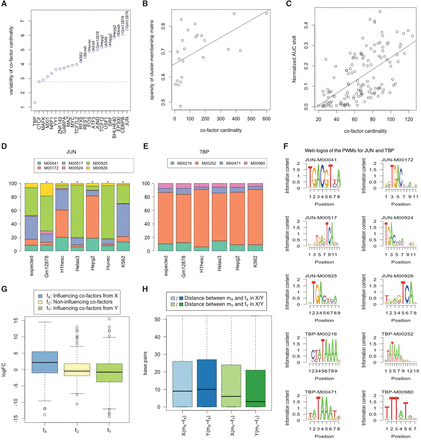
EMT model heterogeneity is associated with cell type specificity of cofactors. (A) The plot shows for each TF the variability of cofactor cardinality across cell types. Each point is labeled by cell type where the relevant TF has specific usage, based on the literature, and has the largest number of cofactors. TBP and CTCF are the most ubiquitous TFs. The green dotted horizontal line denotes the variability of cardinality for CTCF cofactors. (B) Sparsity of cell–membership matrix correlates with cofactor cardinality. (C) Normalized ROC-AUC difference of Interaction and NonInteraction models for a specific TF–cell type pair correlates with cofactor cardinality. (D,E) Motif usage for the reference TF in the NonInteraction models of different cells, for JUN and TBP as two extreme examples. y-axis denotes the feature importance of motif usage in the NonInteraction model. The sequence logos for the corresponding reference PWMs are presented in F. (G,H) fX denotes the influencing cofactors of mX in cell line X; fY, mY in Y. (G, left) Log fold change (logFC) between relevant and nonrelevant cell type for influencing cofactors of mX; (middle) logFC for noninfluencing cofactors; (right) logFC between nonrelevant and relevant cell type for influencing cofactors of mY. (H) Genomic proximity of the motif-specific interaction partner with the motif. mX∼fX denotes the nearest genomic distances (in base pairs) from mX motif to any cofactors in the set of fX and so on.











