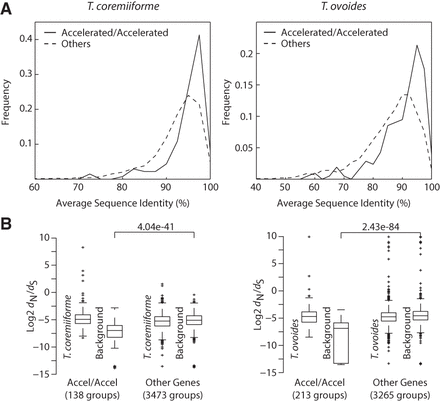
Accelerated evolution of highly conserved genes. (A) Distributions of average amino acid sequence identities taken over all pairwise comparison of genes from species surrounding each hybrid species, i.e., T. coremiiforme–T. asahii–T. faecale gene group or T. ovoides–T. inkin gene group. The comparisons are between gene ortholog groups that became evolutionarily accelerated following a hybridization event (gene groups that are present as two-copy in a hybrid species and both gene copies exhibit acceleration in evolutionary rates) and those that did not (other two-copy genes). Genes with accelerated evolutionary rates are enriched for highly conserved genes. (B) Box plots comparing dN/dS values of hybrid two-copy genes to their corresponding background. The whiskers cover ∼99.3% of the distribution. Data for the evolutionarily accelerated gene group and the rest of the genome are shown. Mann-Whitney U test P-values between the background groups are indicated: (left) T. coremiiforme with the weighted average of T. asahii and T. faecale genes as background; (right) T. ovoides genes with T. inkin genes as background.











