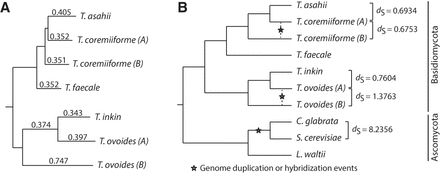
Recent hybridization events in T. coremiiforme and T. ovoides. (A) Phylogenetic tree reconstructed from more than 3000 gene ortholog groups that are present in all nonpolyploid Trichosporon genomes and remain two-copy in either T. coremiiforme or T. ovoides. Subgenome assignments (A and B) were made from (1) gene homeologous relationships, (2) sequence identities to nonhybrid orthologs, and (3) phylogenetic analyses. Subgenome A represents the one that is evolutionarily closer to either T. asahii in the case of T. coremiiforme or T. inkin in the case of T. ovoides. Sequences of genes belonging to each subgenome or genome were concatenated together and used to estimate the average substitution rates. Branch lengths are proportional to the synonymous substitution rates (dS) which are also indicated as branch labels. (B) Phylogenetic relationship between Trichosporon and selected Ascomycota fungi. Pairwise dS estimations were derived based on 822 gene ortholog groups that are present in all species and remain two-copy in both T. coremiiforme and T. ovoides (a total of 10 orthologous/homeologous genes per group). These estimates suggest that the hybridization events in Trichosporon are at least six to 12 times more recent that the genome duplication event in the Saccharomycetaceae lineage.











