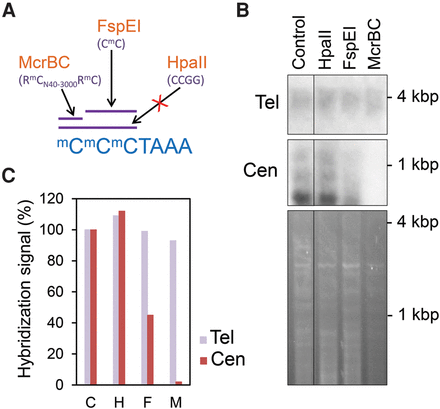
Methylation-dependent restriction enzyme analyses confirm the absence of telomeric DNA methylation. (A) Cartoon representing the sequence specificity of the restriction enzymes used to digest Arabidopsis genomic DNA. (B) Southern blot hybridizations of Arabidopsis genomic DNA digested with the restriction enzymes indicated in A. The upper panel shows equal amounts of undigested (Control) or digested (HpaII, FspEI, or McrBC) DNA samples hybridized with a telomeric probe. The four DNA samples were also digested with Tru9I prior to hybridization. The middle panel shows the hybridization of the same samples with a 180-bp centromeric repeat probe. The ethidium bromide staining of the samples is shown in the lower panel. The control DNA sample was run in the same gel as the rest of the samples, so that their corresponding hybridization signals were processed equally. (C) Bar plot representation of the telomeric and centromeric bottom band hybridization signals expressed as percentages of the control. C, H, F, and M represent Control, HpaII, FspEI, and McrBC, respectively.











