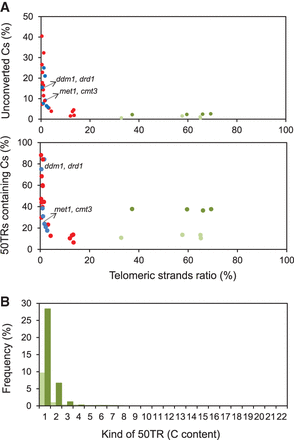
A synthetic nonmethylated telomeric DNA template is not fully converted during bisulfite treatment. (A) Scatter plot representations of the percentages of unconverted cytosines (Cs) within 50TRs and of 50TRs containing cytosines versus the telomeric strands ratio. Ratio values were calculated as the number of 50TRs divided by the number of reads that follow the TTTAGGG pattern along the first 50 bp. They were expressed as percentages. Whole-genome bisulfite sequencing studies performed with the Arabidopsis wild-type strain (red dots) and with DNA methylation mutants (blue dots) were analyzed. Similarly, bisulfite sequencing experiments performed with a 105-bp-long telomeric synthetic fragment at 54°C (light green dots) or at 40°C (dark green dots) were also analyzed. Data corresponding to the met1-3, cmt3-11 and ddm1-2, drd1-7 double mutants are indicated. (B) Bar plot representation showing the distribution of cytosines within 50TRs derived from the synthetic telomeric fragment treated with bisulfite at 54°C (light green bars) or at 40°C (dark green bars). The percentages of 50TRs containing different numbers of cytosines are represented. Average values corresponding to four independent experiments are shown for each temperature.











