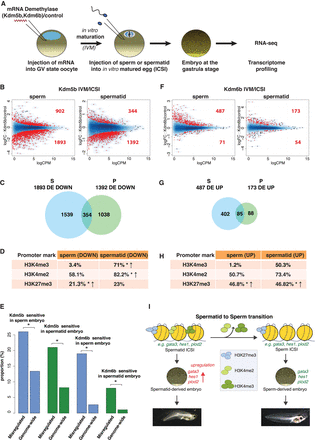
Paternal genome marking by H3K4me2/3 and H3K27me3 is required for gene expression in the embryos. (A) Histone demethylase expression assay. (B) MA plot showing log fold-change (logFC, y-axis) in gene expression between Kdm5b (H3K4me2/3 demethylase)- versus control mRNA-injected embryos, against log counts per million (logCPM, x-axis). Red dots: genes differentially expressed (FDR < 0.05); N = 4 independent experiments. (C) Venn diagram of down-regulated genes upon KDM5B expression in sperm- (blue) and spermatid-derived (green) embryos. (D) Percentages of genes down-regulated upon KDM5B expression in embryos that show H3K4me2/3 and H3K27me3 promoter peaks in the paternal cell. (*) P-value < 0.05 (χ2 test); ↑: over-represented when compared to genome-wide distribution. (E) Proportion of misregulated genes affected in each demethylase expression assay. (*) P-value < 0.05 (χ2 test). (F) Same as B for KDM6B (H3K27me3 demethylase) expression. (G) Same as C with genes up-regulated upon KDM6B expression. (H) Same as D for genes up-regulated upon KDM6B expression. (I) Model of epigenetic programming of sperm for the regulation of embryonic transcription.











