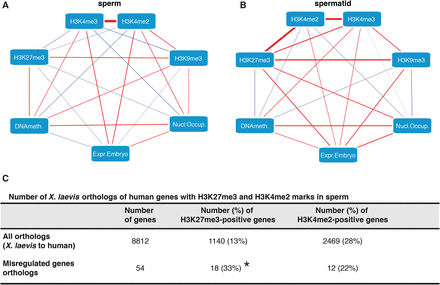
H3K27me3 target genes that lose H3K4me2/3 in sperm compared to spermatids are misregulated in spermatid-derived embryos. (A,B) Differential gene expression between sperm- and spermatid-derived embryos best correlates with differential H3K4me2/3 and H3K27me3 marking in sperm and spermatids. Partial correlation network between all tested epigenetic features of the paternal chromatin (A, sperm; B, spermatid) and gene expression in the corresponding embryos. Edges (lines) represent positive (red) or negative (blue) partial correlations. Edges thickness: strength of the partial correlations. (C) H3K4me2 and H3K27me3 marking on misregulated genes is conserved between Xenopus and human sperm. As compared to all orthologs, the misregulated orthologs are enriched for H3K27me3 marks over the genome-wide average in human sperm (χ2 test, [*] P-value < 0.05). No statistical enrichment for H3K4me2 on misregulated genes as compared to the genome-wide average is observed in human sperm.











