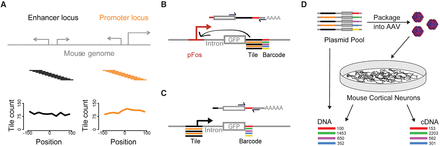
Measuring thousands of promoter and enhancer activities in primary neurons using Massively Parallel Reporter Assays (MPRAs). (A) For each of 234 CREBBP-bound promoter and 253 CREBBP-bound distal enhancer loci, we synthesized 11 139-bp tiles whose start sites are spaced at 20-bp intervals. The center tile at each locus is centered on the called CREBBP peak. Plots show tile coverage as a count of quantifiable tiles versus position relative to the locus center. Distal enhancers are at least 500 bp from an annotated TSS. (B) After a series of cloning steps (Supplemental Figs. S1, S2), we tagged each tile with one or more unique 18-bp barcodes. In the enhancer test, the abundance of each barcode in a cellular mRNA pool depends on the enhancer activity of its associated tile. The library contains a mix of tiles from enhancer (black) and promoter (orange) loci. pFos is a 100-bp basal FOS promoter. (C) In the promoter test, the abundance of each barcode depends on the promoter activity of its tile. (D) We packaged MPRA libraries into AAV for infection of mouse cortical neurons and quantified cDNA and control DNA barcode abundance by sequencing. We typically sequenced control DNA amplified from the plasmid library rather than from cells, because we found MPRA read counts from plasmid DNA and DNA extracted from neurons to be indistinguishable (Supplemental Fig. S3C).











