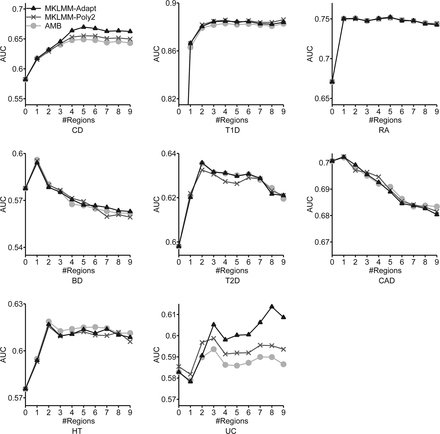
Evaluation of prediction performance in WTCCC data sets as a function of the number of selected regions: (CD) Crohn's disease; (T1D) type 1 diabetes; (BD) bipolar disorder; (RA) rheumatoid arthritis; (T2D) type 2 diabetes; (CAD) coronary artery disease; (HT) hypertension; (UC) ulcerative colitis. The AUC for T1D with zero regions is 0.589 for all methods and is omitted for clarity. GBLUP is not shown explicitly, because AMB with no selected regions is equivalent to GBLUP. MKLMM-Adapt performed as well as or better than AMB across all data sets (evaluated at the number of regions at which prediction performance peaked). Prediction performance always peaked at a certain number of regions and then dropped, indicating that the models may overfit the data when an overly large number of regions is selected. The phenotypes where MKLMM-Adapt performed significantly better than AMB (CD, T1D, UC) appear to be those in which many regions are required for good performance, implying a complex genetic architecture.











