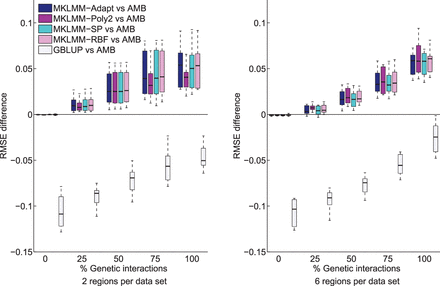
Comparison of the evaluated methods on synthetic data sets with various levels of genetic interactions and numbers of genomic regions harboring interacting variants. The box plots show the difference between the evaluated methods and Adaptive MultiBLUP (AMB), in terms of the root mean square error (RMSE) between the predicted and observed phenotype. Larger values indicate a greater advantage over AMB. The advantage of the MKLMM methods over AMB increased with the percentage of genetic interactions. MKLMM-Poly2 gains a slight advantage over the other MKLMM methods in the presence of six regions, because its simpler model is less prone to overfitting when many parameters need to be estimated from the data.











