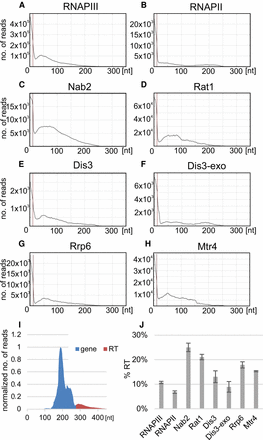
Genome-wide analysis of surveillance factors and its role in targeting transcriptional read-through of tRNA genes. Genome-wide profile of nuclear tRNA transcripts aligned to mature 3′-ends. Genome-wide hit distributions are shown for (A) RNAPIII (Rpo31), (B) RNAPII (Rpo21) (Holmes et al. 2015), (C) Nab2, (D) Rat1 (Granneman et al. 2011), (E) Dis3, (F) Dis3-exo, (G) Rrp6, and (H) Mtr4. (I) Schematic representation of genome-wide profile divided into gene and RT fragments. The frequency of binding to RT regions was calculated as RT fraction divided by total number of reads (gene plus RT). (J) Histogram showing frequency of binding to RT regions (in percentages) relative to total tRNA for genome-wide profiles.











