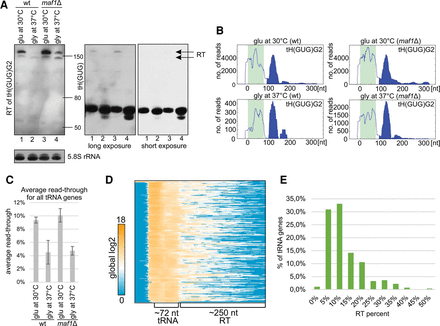
Transcription termination read-through (RT) by RNAPIII. (A) Northern blotting reveals extended transcripts from gene tH(GUG)G2 (left). Total RNA extracts were analyzed by Northern blotting using probes specific to the RT sequence 54 nt downstream from the tRNA (left), within tH(GUG) (right), and 5.8S rRNA (loading control). Extended forms are of low abundance relative to mature tH(GUG). (B) Distribution of read density over gene tH(GUG)G2 under conditions presented in A. These CRAC results are consistent with the Northern blot but indicate the instability of extended transcripts. (C) Average RT levels for all nuclear tRNA genes. RT was calculated for each gene under indicated conditions separately, and average RT values for all nuclear tRNA genes are presented. (D) Heatmap for all nuclear tRNA genes, with 50-nt 5′ flanks and 250-nt 3′ flanks, clustered using the 1-Pearson coefficient. Hits intensity is presented as log2 of global values. Regions of mature tRNA genes and RT are labeled. (E) Histogram showing distribution of RT among all tRNA genes.











