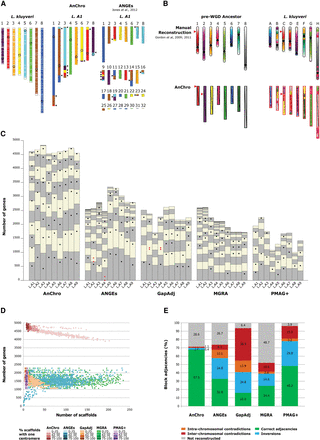
(A) Comparison between the two versions of the L.A1 ancestral genome reconstructed by AnChro and by ANGES (Jones et al. 2012). Chromosome painting representations of ancestral genomes are colored relatively to the L. kluyveri chromosomes. The black triangles indicate the same 12 ancestral adjacencies that resulted from six translocations identically reconstructed by the two tools. (B) Comparison between the manually reconstructed (Gordon et al. 2009, 2011) and the AnChro version of the pre-WGD ancestral genome relative to the L. kluyveri genome. The only inter-chromosomal difference between the two reconstructions is indicated by the red triangles. (C) Comparison of the nine Lachancea ancestors (L.A1 to L.A9) reconstructed by AnChro, ANGES, GapAdj, MGRA, and PMAG+. Synteny blocks were computed with I-ADHoRe for the five reconstruction tools. For AnChro, a single default reconstruction is presented. Each column represents the ancestral chromosomes of a given ancestor as an alternation of gray and beige boxes, with size being proportional to the number of reconstructed ancestral genes. The small black circles indicate the centromere position. The small red circles indicate the centromere positions when an ancestral chromosome was reconstructed with two centromeres. (D) Ancestral genome reconstructions on simulated genomes. The figure presents 900 reconstructed ancestral genomes corresponding to nine different ancestors per simulation and 100 simulations, performed with AnChro (default reconstructions), ANGES, GapAdj, MGRA, and PMAG+. Each genome reconstruction is represented by a dot. The quality of the reconstructions is assessed by three criteria: the number of ancestral scaffolds (ideally eight), the number of ancestral genes (ideally 5000), and the proportion of scaffolds per reconstruction with a single centromere (ideally 100%). (E) Average proportions of correctly and incorrectly reconstructed adjacencies for the 900 reconstructions obtained by the five tools. Incorrect adjacencies are decomposed in single block inversions and intra- and inter-chromosomal contradictions. The average proportion of adjacencies that were not reconstructed by the different software is also indicated.











