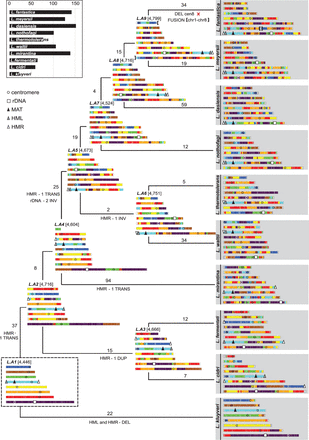
Chromosomal history of the Lachancea genomes. The chromosomal structures of the 10 extant species and the ancestral genomes L.A2 to L.A9 are represented as a function of the genome structure of L.A1, the last common ancestor of the clade. The number of genes of each ancestral genome is indicated with brackets. The total number of translocations and inversions accumulated between two genomes is indicated above each branch. Rearrangements involving MAT, HML, HMR, rDNA, or centromeres are indicated below each branch. The relocation of the rDNA array occurred in the branch between L.A4 and L.A5. This transposition event occurred intra-chromosomally from an ancestral site, represented as a purple region in L.A4, to a new genomic location close to the green centromere in L.A5. The relative orientation of the rDNA and the centromere was inverted between L.A4 and L.A5, suggesting that the rDNA relocation resulted from two intra-chromosomal inversions involving one breakpoint reuse (Supplemental Fig. S5; Supplemental Table S6). The interval between MAT and HML was never broken and was inherited intact from L.A1 in all extant species except L. kluyveri, which lost both HML and HMR. The HMR cassette underwent many rearrangements (three translocations, one inversion, and one duplication) but always remained subtelomeric. The HML, HMR, and the MAT loci were located on the same chromosome in the last common ancestor of the genus, L.A1, with one silent cassette at each chromosome end. The only Lachancea species that also harbors the three sexual loci on a single chromosome is L. fermentati, but this organization is not inherited from L.A1 as it results from additional translocations in the branch between L.A3 and L. fermentati. Therefore, none of the present-day Lachancea species has retained the original chromosomal organization of the sexual loci. The inset plot recapitulates the total number of translocations and inversions that accumulated since each extant species diverged from the last common ancestor, L.A1.











