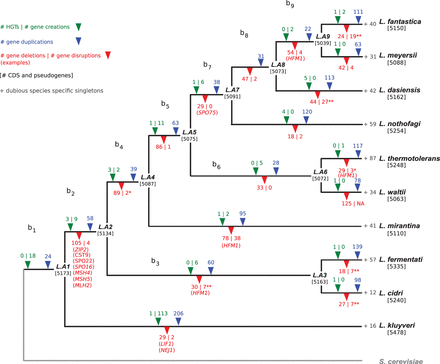
Evolution of the Lachancea gene repertoire. The number of gene duplications (in blue) and losses (in red) were estimated on each branch of the tree under a birth–death evolutionary model (Methods). The total number of gene losses indicated on the figure (1036) comprises the 107 cases of dubious loss (see text). The statistical significance of the enrichment of gene losses associated to rearrangements compared to the proportion of genes associated to rearrangements is indicated by an asterisk (P < 0.05) or double asterisk (P < 10−4). Notable examples of gene losses are in parentheses below their corresponding branches. The phyletic patterns of the 51,110 CDS and 1018 pseudogenes were used to map HGTs and gene creation events (in green) in the different branches of the tree (Methods). The number of species-specific singletons is in gray at the tip of each terminal branch. The total number of genes in each ancestral genome and the number of genes and pseudogenes in extant species are indicated between squared brackets.











