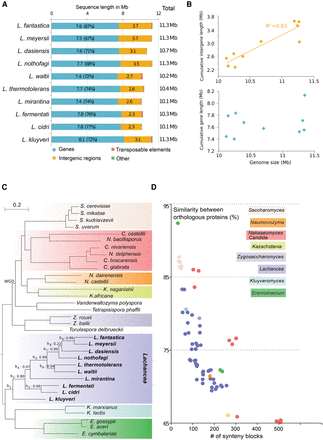
(A) Cumulative sequence length of the annotated genetic elements in the 10 Lachancea genomes. The percentages of protein-coding sequences are in parentheses. (B) Genome size in Lachancea positively correlates with intergene length (top) but not with cumulative gene length (bottom). (C) Phylogeny of 34 Saccharomycetaceae species inferred from a maximum likelihood analysis of a concatenated alignment of 756 families of syntenic homologs. The tree topology within the Lachancea clade remains identical for several reconstruction methods: concatenation tree, majority tree, and extended majority rule consensus (eMRC) tree (see Methods). Internal branches within the Lachancea clade are named b1 to b9. The corresponding internode certainty (IC) values, indicating the robustness of the eMRC tree topology, are given. (WGD) Whole-genome duplication. (D) Relationship between orthologous protein similarity and the number of conserved synteny blocks within different yeast genera. The Lachancea genus is the only clade showing a continuum of genome reorganization and pairwise protein similarities over a large evolutionary range.











