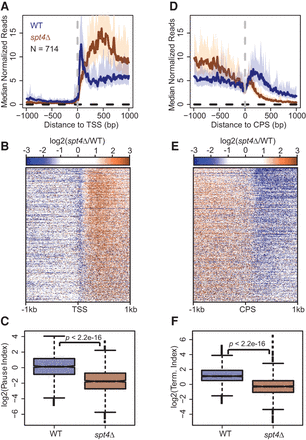
5′ and 3′ ends of genes exhibit a loss of Pol II density as a result of Spt4 deletion in S. pombe. (A,D) Median PRO-seq signal around the observed TSS (A) or CPS (D) of active, filtered genes longer than 1 kb and separated from the boundaries of neighboring genes on the same strand by at least 1 kb. Medians reflect 10-bp bins, and the 12.5% and 87.5% quantiles are shown in light shaded regions. (B,E) Heatmaps of log2 fold change of mutant versus wild type per 10-bp bin around the TSS (B) or CPS (E) for all genes used in A and D. Genes within heatmaps are sorted by decreasing amount of wild-type PRO-seq reads within the first 500 bp downstream from the TSS. (C) Box plots showing the distribution of pausing index values for WT and spt4Δ in S. pombe. (F) Box plots showing the distribution of termination index values for WT and spt4Δ in S. pombe. P-values represent the results of Student's t-test.











