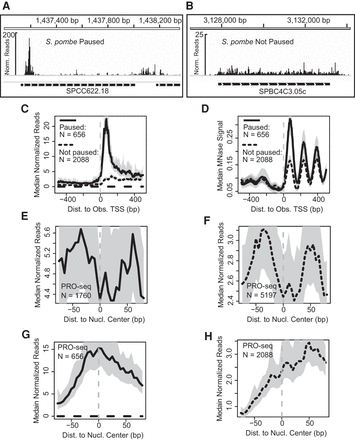
Pol II distributions at paused genes in S. pombe are coupled with increased nucleosome occupancy or positioning. (A,B) Browser images of PRO-seq read counts across S. pombe genes classified as either high-confidence paused (A) or not paused (B). (C) Median PRO-seq read counts within 1 kb upstream of and downstream from the observed TSS of paused and not paused genes. (D) Median MNase-seq read coverage within 1 kb upstream of and downstream from the observed TSS of paused and not paused genes. (E,F) PRO-seq signal around gene-body nucleosome centers for paused (E) and not paused genes (F). (G,H) PRO-seq signal around +1 nucleosome centers for paused (G) and not paused genes (H). For metagene plots in C–H, medians reflect 5-bp bins, and the 12.5% and 87.5% quantiles are shown as shaded regions. All panels represent profiles of combined wild-type biological replicates.











