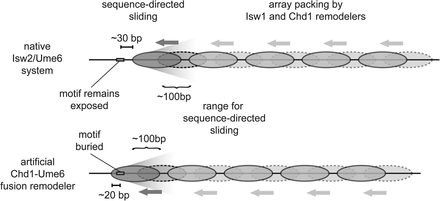
Model for transcription factor-mediated chromatin remodeling at sequence-targeted sites. (Top) In the endogenous system, Isw2 is recruited to specific genomic loci through interaction with Ume6 at target motifs (rectangle). Motif-proximal nucleosomes (dark gray) within a limited range are shifted by Isw2 from an unremodeled position (dashed lines) to a final position such that 30 bp of DNA are exposed between the motif center and the nucleosome edge. (Bottom) The artificial Chd1-Ume6 system works through a similar mechanism where the motif proximal nucleosome is directly moved by the chromatin remodeling protein, but with this remodeler, the recruitment motif is buried by 20 bp of nucleosomal DNA. For both cases of sequence-directed nucleosome sliding, the motif-proximal nucleosome determines the phasing for evenly spaced nucleosome arrays that are packed against these founding nucleosomes by endogenous remodelers.











