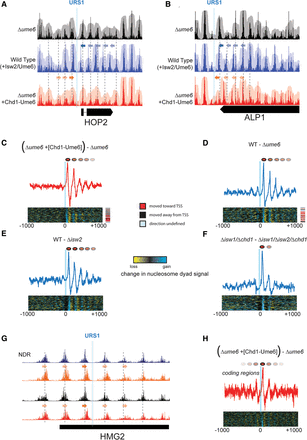
Positioning of motif-proximal nucleosomes sets the phase for nucleosome arrays. (A) Nucleosome positioning by Isw2/Ume6 (blue) and Chd1-Ume6 (red) at the HOP2 locus compared with lack of Ume6-directed positioning (black). Solid arrows indicate the direction of movement for motif-proximal nucleosomes, while faded arrows denote secondary shifts in nucleosome positions. Gray dashed lines indicate preferred nucleosome dyad positions in the Δume6 strain. (B) Same as in A for a Ume6 motif at the 3′ end of the ALP1 gene. (C) Difference in nucleosome dyad signal for Δume6 strains with and without Chd1-Ume6 for motif-proximal and distal nucleosomes within 1000 bp of the Ume6 binding motif. Signal is shown for motifs corresponding to cluster 3 (right-to-left repositioning), though cluster 1 displayed similar results. (Top) Average difference in nucleosome dyad signal within the cluster; (bottom) difference in signal at all individual motif instances within the cluster. Direction of nucleosome positioning relative to associated TSS (if applicable) is indicated on right. (D–F) Same as in C, but for differences between (D) the wild type and Δume6, (E) the wild type and Δisw2, or (F) Δisw1/Δchd1 and Δisw1/Δchd1/Δisw2 (Gkikopoulos et al. 2011). (G) Example of nucleosome boundary formation within a coding region, annotated as in A and B. (H) Average difference in nucleosome dyad signal as in C for 77 instances of coding region nucleosome positioning by Chd1-Ume6.











