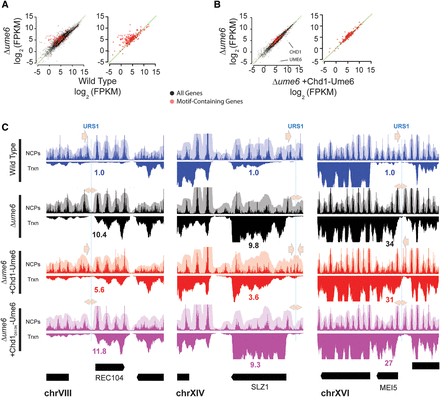
Nucleosome positioning by Chd1-Ume6 is mildly repressive. (A) Differences in transcription between the wild type and Δume6 at all genes (black) or at genes with a WNGGCGGCWW motif within 500 bp of the open reading frame (red). Transcript abundance was calculated using Cufflinks (Trapnell et al. 2012). (FPKM) Fragments per kilobase of transcript per million fragments mapped. Green diagonal indicates the y = x axis. Points above the diagonal are repressed in wild-type cells. (B) Differences in transcription (log2 FPKM) in Δume6 cells with or without addition of Chd1-Ume6 at all genes (black) or genes with associated WNGGCGGCWW motifs (red). Green diagonal indicates the y = x axis. Points above the diagonal are repressed upon introduction of Chd1-Ume6. (C) Example loci showing nucleosome positioning (upper panels), RNA signal (lower panels), and relative transcript abundance from RNA-seq data (associated numbers) at indicated genes for the wild-type, Δume6, and Δume6 + [Chd1-Ume6] and Δume6 + [Chd1D513N-Ume6] strains. Blue lines represent associated Ume6 recognition motifs, and arrows denote directional closing (single-headed arrow) or opening (double-headed arrow) of associated NDRs.











