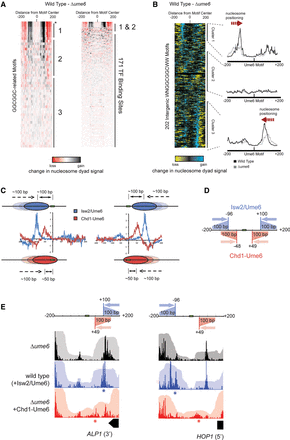
Genome-wide nucleosome positioning by Isw2/Ume6 at Ume6 target sequences. (A) Difference in nucleosome dyad signal at permutations of Ume6 recognition motifs for wild-type and Δume6 strains as in Figure 2B. Black and red indicate where remodeling by endogenous Isw2/Ume6 caused increases and decreases in nucleosome signal, respectively. Asterisk indicates WNGGCGGCWW motif. Ordering is the same as in Figure 2B. (B) The difference in nucleosome dyad signal between wild-type and Δume6 strains at all intergenic instances of the WNGGCGGCWW motif, clustered by direction of nucleosome repositioning (left) with average dyad signal within each cluster for each strain (right) as in Figure 2C. (C) Comparison of nucleosome positioning by Chd1-Ume6 and endogenous Isw2/Ume6 at Ume6 binding motifs. Each trace reveals the average sites of gain (positive) or loss (negative) of nucleosome dyad signal when Δume6 is compared with Chd1-Ume6 (red) or Isw2/Ume6 (blue) at Ume6 binding motifs from clusters 1 (left) or 3 (right). (D) Schematic representation of the range and direction of nucleosome positioning for Isw2/Ume6 (blue) or Chd1-Ume6 (red) at Ume6 binding sites. In comparing the wild type with the Δume6 or Δisw2 backgrounds, nucleosomes situated up to 200 bp from the Ume6 recognition sequence in a Δume6 or Δisw2 strain were shifted by Isw2 to a final dyad position ∼100 bp from the recognition motif. Likewise, nucleosomes with dyads situated up to 150 bp from the motif center in a Δume6 or Δisw2 background were shifted by Chd1-Ume6 to a final dyad position ∼50 bp from the motif (corresponding to burial of the motif by ∼20 bp). (E) Example loci demonstrating differences in nucleosome distributions for Isw2/Ume6 or Chd1-Ume6 in wild-type or Δume6 +[Chd1-Ume6] strains compared with a Δume6 strain. Nucleosome dyad signal (opaque) and associated nucleosome footprints (transparent) are shown for each strain with the predicted nucleosome movements by Isw2/Ume6 (blue) or Chd1-Ume6 (red) shown above, and the expected final dyad positions are represented by asterisks.











