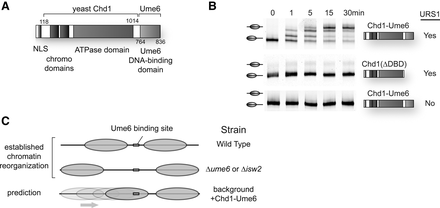
Strategy for targeting of chromatin remodeling activity in vivo. (A) Schematic of Chd1-Ume6 fusion protein. A fragment of S. cerevisiae Chd1 (residues 118–1014) was fused to the DNA binding domain of S. cerevisiae repressor Ume6 (residues 764–836) and cloned into the p416-ADH expression vector (Mumberg et al. 1995) for stable yeast transformation. (B) Sequence-directed nucleosome sliding by Chd1-Ume6 in vitro. Nucleosomes (50 nM) containing a URS1 site (TGGCGGCT) 8 bp from the edge of an end-positioned, FAM-labeled mononucleosome with 80 bp of extranucleosomal DNA were incubated with 10 nM of Chd1-Ume6 (top) or Chd1118-1014 lacking a DNA binding domain (middle) for the indicated times, and nucleosome positions were separated by 6% native PAGE. Under these conditions, nucleosome sliding required both the Ume6 DNA binding domain (middle) and the URS1 binding site (bottom). (C) Summary of strains used and expected outcomes for chromatin remodeling by Chd1-Ume6 on transcription and nucleosome positions at genomic target sequences.











