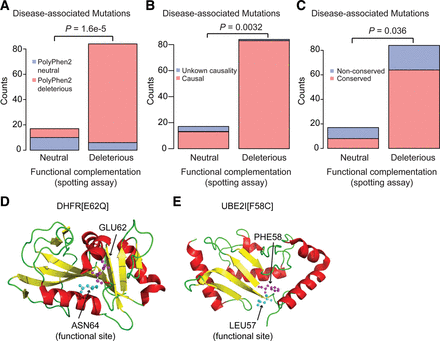
Exploring differences between functional assays and current pathogenicity annotation. (A–C) Among variants currently annotated as disease-associated, those classified as neutral by the FCS assay (apparent false negatives) overlap significantly with (A) variants that are also classified as neutral by PolyPhen2 (P = 1.6 × 10−5, Fisher's exact test); (B) disease-associated variants with unknown causality (P = 0.0032, Fisher's exact test); and (C) variants located at nonconserved residue positions. Enrichment in each case was relative to the corresponding frequency among variants called deleterious by the FCS assay (P = 0.036, Fisher's exact test). (D,E) Among non-disease-associated variants classified as deleterious by functional assays (apparent false positives), there were two variants for which structural context supported the finding of these variants to be deleterious: (D) DHFR[E62Q] and (E) UBE2I[F58C]. (Pink) Residues where variants occur; (cyan) functional sites.











