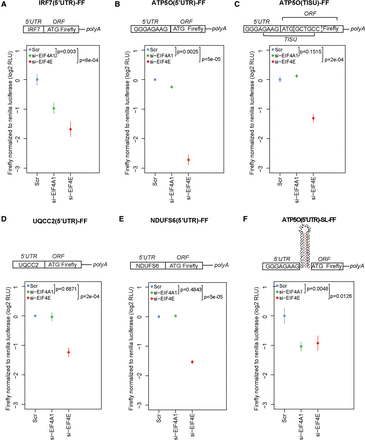
Translation of reporter mRNAs harboring short 5′ UTRs is sensitive to EIF4E, but not EIF4A1. HEK293E cells were transfected with a control (Scr), EIF4E (si-EIF4E), or EIF4A1 (si-EIF4A1) siRNAs and then transfected with firefly (FF) reporters harboring IRF7 5′ UTR (IRF7[5′UTR]-FF) (A), ATP5O 5′ UTR with a proximal portion of TISU element before the initiation codon (ATP5O[5′UTR]-FF) (B), ATP5O 5′ UTR with a full TISU element (ATP5O[TISU]-FF) (C), UQCC2 5′ UTR (UQCC2[5′UTR]-FF) (D), NDUFS6 5′ UTR (NDUFS6[5′UTR]-FF) (E), or ATP5O 5′ UTR followed by a stem–loop structure (ATP5O[5′UTR]-SL-FF) (F). As a control, cells were cotransfected with a Renilla reporter. Luminescence was monitored 48 h post-transfection. Data for firefly luminescence normalized to Renilla luminescence were log2 transformed and normalized per replicate and to the mean of Scr (see Supplemental Fig. 14). Each experiment was performed in independent triplicates, each consisting of three replicates, and data are shown as mean ± SD. RLU, relative light units. Schematics of the FF constructs are provided in the above histograms, and the positions of TISU and ORF of FF luciferase are indicated. P-values from one-way ANOVAs are shown.











