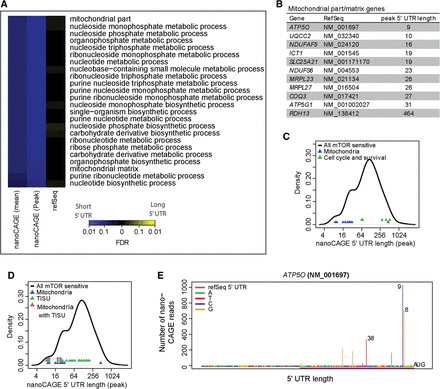
Mitochondria-related transcripts are enriched among MTOR-sensitive mRNAs with extremely short 5′ UTRs. (A) Enrichment of biological processes in MTOR-sensitive mRNAs (from polysome-profiling) depending on nanoCAGE or RefSeq 5′ UTR lengths. Colors represent false discovery rates for enrichments among short (blue) or long (yellow) 5′ UTRs. Both mean and largest peak 5′ UTR lengths from nanoCAGE were used as input. (B) A table of genes from the mitochondria-related functions (identified in A) and their peak 5′ UTR lengths as determined by nanoCAGE. (C) A comparison of 5′ UTR lengths of MTOR-sensitive mitochondrial function or cell cycle/survival related genes. (D) Comparison of 5′ UTR lengths and the presence of a TISU element in the indicated subsets of MTOR-sensitive mRNAs. (E) ATP5O is a subunit of complex V of the electron transport chain (ATP synthase) and is encoded by mRNA with a putative TISU motif and an extremely short 5′ UTR (plot as in Fig. 2C).











