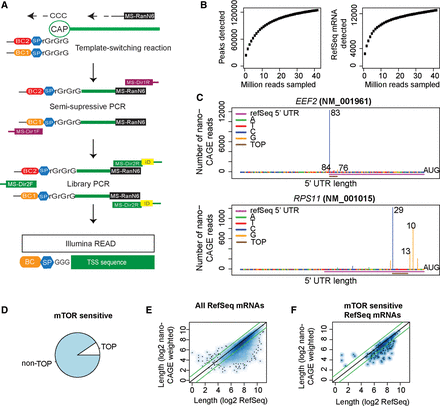
Transcription start site analysis using nanoCAGE confirms that MTOR regulates translation of non-TOP mRNAs. (A) The nanoCAGE procedure to identify transcription start sites (TSS). (BC, barcode; SP, spacer; MS-RanN6, random hexamer; MS-Dir1R/F, primers for semisuppressive PCR; MS-Dir2R/F, primers for adaptor PCR). (B) A comparison of the number of identified peaks (more than five reads; left) and RefSeq mRNAs (>50 reads; right) under increasing number of sampled nanoCAGE sequencing reads. (C) TSS revealed by nanoCAGE in two TOP mRNAs. The UTR lengths suggested by the three major TSS peaks are indicated together with the 5′ UTR region according to RefSeq and the position of the 5′ TOP motif. (D) The proportion of MTOR-sensitive (by polysome-profiling) mRNAs with TOP elements according to nanoCAGE. (E,F) A comparison between 5′ UTR lengths according to RefSeq and nanoCAGE mean lengths for all RefSeq mRNAs detected (E) and MTOR-sensitive genes (F). Green lines indicate a twofold difference in 5′ UTR length.











