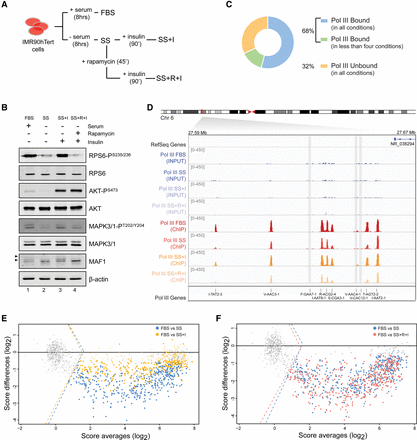
Genome-wide analysis of Pol III recruitment reveals differential occupancy profiles under prosurvival and stress conditions. (A,B) IMR90hTert cells were grown in the presence (FBS) or absence (serum-starved, SS) of fetal bovine serum for 8 h. We then added 1 µM insulin to starved cells alone (SS + I) or after treatment with 2 nM rapamycin (SS + R + I), for the indicated times. Whole-cell extracts were subsequently analyzed by immunoblotting for total levels and phosphorylation status of the indicated proteins. The MAF1 hyperphosphorylated forms are indicated by arrows. (C) Pie chart representation of 715 Pol III genes either unbound (226) or bound (489) to polymerase in at least one of the four conditions (FBS, SS, SS + I, or SS + R + I). (D) Pol III occupancy as measured by ChIP-seq, shown for a cluster of tRNA genes on Chromosome 6. Chromatin was prepared from IMR90hTert cells grown as described in A and immunoprecipitated with an antibody against the POLR3D (RPC4) subunit of Pol III. Input DNA signal is shown. Gray shadings indicate nonoccupied genes. tRNA gene names are according to HGNC-approved gene nomenclature (“TR” in front of names is omitted for space constraints). (E,F) MA-plots showing distributions of Pol III occupancy scores [log2 (IP/input)] for 715 loci. In each panel, two comparisons are superimposed: SS versus FBS (blue) and SS + I versus FBS (yellow) in E; SS versus FBS (blue) and SS + R + I versus FBS (pink) in F. Gray dots left of the dotted lines (indicating the cutoff threshold score) represent nonoccupied genes. Small dots are genes that do not change significantly (P > 0.005) in each pairwise comparison. The horizontal line indicates the zero-fold change.











