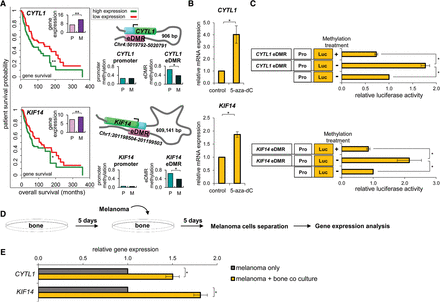
Melanoma coculture with osteoblasts alters pro-cancer eDMR-associated gene expression. (A) Schematic representation of the eDMR-gene pairs: Chr4:5019792–5020791 for CYTL1 and Chr1:201198504–201199503 for KIF14. The eDMR (pink) is interacting with the promoter (cyan) of the gene (green) through chromatin looping. Kaplan–Meier survival plots (left panels) show significant differences between patient outcomes at high and low expression levels (green and red curves, respectively) of CYTL1 and KIF14 genes. (Insets) Bar graphs show up-regulation of CYTL1 and KIF14 in melanoma bone metastases. Middle panels: Methylation of the promoters of these genes does not vary between primary melanoma (P) and melanoma bone metastases (M). Right panels: Enhancers are differentially hypomethylated in melanoma bone metastases (Wilcoxon rank-sum tests, [*] q < 0.15, [**] q < 0.05; FDR adjusted). (B) WM3682 melanoma cells were treated with 10 µm 5-aza-dC or with DMSO (control) followed by quantification of CYTL1 and KIF14 mRNA levels that were normalized to GAPDH. Data are relative to levels in control-treated cells. Error bars represent ± SEM; (*) P < 0.05; N = 3. (C) WM3682 melanoma cells were transfected with methylated CYTL1-eDMR or KIF14-eDMR reporter plasmids, unmethylated eDMR reporter, or reporter without eDMR (control). Firefly luciferase activity is normalized to Renilla luciferase. Fold-changes are relative to control. Error bars represent ± SEM; (*) P < 0.05; N = 3. (D) Experimental design scheme. (E) Levels of CYTL1 and KIF14 mRNA were determined in melanoma cells before and after coculturing with osteoblasts. Data were normalized to levels of actin. Error bars represent ± SEM; (*) P < 0.05; N = 3.











