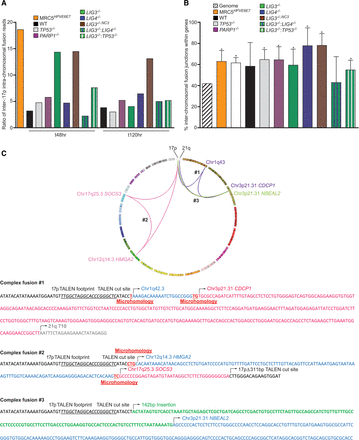
Characteristics of inter-chromosomal fusions between 17p and diverse genomic loci sequenced from MRC5HPVE6E7 cells undergoing telomere-driven crisis or HCT116 cells undergoing 17p TALEN-induced DNA damage and repair. (A) Ratios of Illumina HiSeq 2000 sequence reads mapping to inter-chromosomal telomere-genomic linkages compared with 17p–17p intra-chromosomal linkages are plotted to display enrichment of inter-chromosomal links for all samples. Results relating to specific DNA repair-deficient HCT116 lines are separated by the colors expressed in the key. (B) Inter-chromosomal fusion junctions for all samples are more frequently coincident with genes—analysis restricted to Ensembl (Cunningham et al. 2015) and RefSeq (Pruitt et al. 2014) curations—than expected by chance based on RefSeq human genome gene content estimate of 41.8%. Junction locations were investigated using the UCSC Genome Browser (Kent et al. 2002), and statistical significance was determined by χ2 analysis for all samples, except HCT116 WT and LIG3−/−:LIG4−/− cells: (*) P < 0.05. (C) Telomere-genomic fusion events have the potential to be permissive for cellular transformation via telomerase reactivation. A Circos (Krzywinski et al. 2009) plot illustrating three independent inter-chromosomal fusion events between TALEN-targeted 17p and genomic loci implicated in TERT regulation—Chr3p21.31 (Cuthbert et al. 1999) and Chr12q14.3;HMGA2 (Li et al. 2011)—identified by Illumina HiSeq and Sanger sequencing of fusion amplicons from HCT116 PARP1−/− (#1 and #2, purple and pink linkages) or LIG3−/− (#3, green linkages) cells. The junction sequences of these linkages are presented below the Circos plot, with TALEN recognition and cleavage sites, insertions, and microhomology features indicated.











