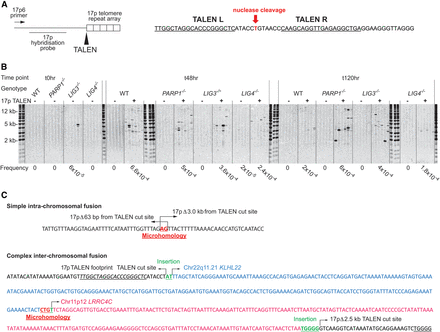
Development of targeted nucleases to induce DSBs and fusions at the 17p telomere. (A, left) Schematic depicting the subtelomeric region of the 17p Chromosome arm with TALEN target site at the start of the telomere repeat arrays indicated (arrow). The 17p6 primer used to amplify and the location of the 17p hybridization probe used to detect 17p fusion events are shown. (Right) The specific recognition sequences bound by the left (L) and right (R) TALEN nucleases to create a functional heterodimer that cleaves the intervening T residue juxtaposing the 17p telomere hexameric TTAGGG repeats. (B) Nucleofection of HCT116 cell lines with the 17p TALEN pair (+) resulted in diverse amplifiable 17p telomere fusions that were absent from untransfected cells (−) at 48 and 120 h post-nucleofection. A sequence-characterized 17p amplicon stochastically detected in the pretransfected (t0) HCT116 LIG3−/− cells is shown. Telomere fusions were detected following Southern blotting using the 17p hybridization probe. Fusion frequency estimated empirically is recorded beneath the panels. (C) Examples of simple intra-chromosomal (upper) and complex inter-chromosomal (lower) 17p TALEN-induced telomere fusions characterized by Sanger sequencing of reamplified and purified fusion amplicons. The specificity of 17p TALEN targeting is indicated by the proximity of fusion junctions to the TALEN cleavage site in one chromatid: (Δ) deletion from this position. The TALEN recognition site is in italics and underlined. Junction insertions and microhomology are marked above and below the sequence, respectively. The two distinct loci incorporated are separated by color and annotated with their genomic locations.











