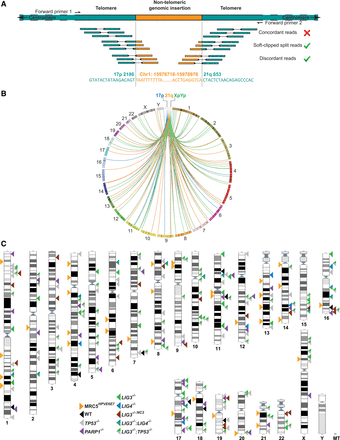
Characterization of telomere-genomic inter-chromosomal fusions in crisis-stage MRC5HPVE6E7 cells. (A) Cartoon representation of an inter-chromosomal fusion between the 17p and 21q telomeres (green) and a genomic locus (orange) amplified from MRC5HPVE6E7 fibroblasts undergoing telomere-induced crisis and sequenced by Illumina HiSeq 2000 paired-end sequencing. Discordant read pairs are those that do not map to the reference sequence with the expected orientation or size coverage. Soft-clipped reads are those containing mismatches with the reference sequence. Inter-chromosomal telomere-genomic fusion events were defined as those discordant and soft-clipped linkages mapping to at least one telomere end (17p, XpYp, or the 21q family) and a nontelomeric genomic location. Fusion PCR primer orientations are indicated above the chromatids. (B) Circos plots (Krzywinski et al. 2009) displaying all inter-chromosomal fusion linkages between the 17p, XpYp, and 21q family telomeres and genomic loci sequenced from crisis-stage MRC5HPVE6E7 cells. A scaled representation of each human chromosome in clockwise orientation with numerical identifiers creates the circumference of the plot. The particular telomeres investigated in this study are featured as separate references at the top of the plot. Linkages between the genome and each telomere are distinguished by the color of the lines traversing the plot. (C) Karyotype map showing coordinates of all sequence-verified and BLAST-authenticated telomere-genomic inter-chromosomal fusion junctions identified in crisis-stage MRC5HPVE6E7 cells (arrows on the left of the chromosomes) and HCT116 cell lines (arrows on the right of the chromosomes) selectively compromised in components of DNA repair pathways and subjected to TALEN-targeted nuclease-induced DSBs at the 17p telomere. Each arrow represents a sequenced fusion junction between a telomere and the genome, and the different samples are distinguished by color, as indicated in the key.











