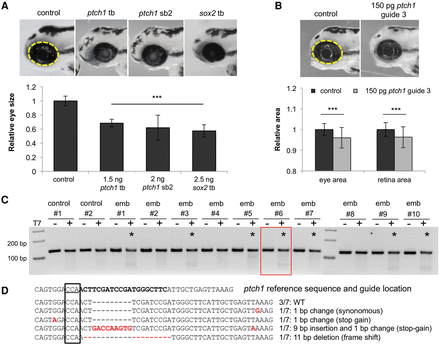
Suppression of ptch1 in vivo results in microphthalmia. (A) Representative lateral images of uninjected control and ptch1 tb, ptch1 sb2, or sox2 MO-injected embryos at 3 d post-fertilization (dpf) (top). Dashed yellow line indicates area of measurement used for quantification of eye size; morphant measurements were normalized relative to control (bottom). (B) Representative lateral images of uninjected control and ptch1 guide RNA (gRNA)/Cas9 injected embryos at 3 dpf (top). ptch1 CRISPR F0 embryos were scored for eye size and retina size (bottom). (C) At 1 dpf, a representative sampling of 10 founders and two uninjected controls were selected and subjected to T7 endonuclease 1 (T7E1) assay. The appearance of T7E1 fragments < 100 bp (marked by asterisks) indicate positive gRNA targeting of exon 6 in the ptch1 locus. No T7E1 fragments were detected in uninjected control embryos. Of the 10 founders subjected to T7E1 assay, seven showed the presence of T7E1 fragments, indicating that ∼70% of founders have insertion/deletions (indels) in the exon 6 region of ptch1. (D) Multiple sequence alignment of ptch1 reference sequence to ptch1-CRISPR variants generated from PCR amplification, subsequent TA cloning, and sequencing of ptch1-gRNA/Cas9 injected embryo #6. Black bold font marks the guide target, and the PAM recognition motif is underlined. Seven PCR-cloned sequences are shown, representing three wild-type variants and all four changes detected. TA cloning and sequencing of each founder embryo indicated 10%–65% mosaicism in individual fish (n = 5 assessed).











