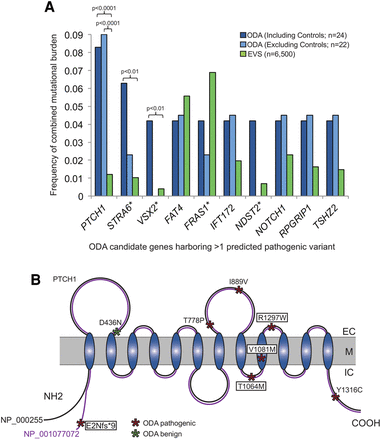
PTCH1 has a significantly enriched mutational burden in ocular developmental anomalies (ODA). (A) Mutational burden for all genes harboring >1 rare predicted pathogenic variants in the initial cohort of ODA compared to healthy control individuals from the Exome Variant Server (EVS). Frequency of combined mutational burden is shown for the 10 genes harboring multiple rare (<1% alternate allele frequency) functional variants (frameshift, nonsense, and splicing variants were considered as damaging; missense variants were classified as damaging or not based on PolyPhen-2) in either ODA cases including two positive controls (n = 22 unknown + 2 positive controls; dark blue bars), or ODA test cases alone (n = 22 unknown; light blue bars) vs. EVS controls (n = 6500; green bars). P-values are indicated for the only three genes with a significant enrichment in cases versus controls (χ2 test). (*) VSX2 and STRA6 are previously identified causal genes in the positive control ODA samples, C1 and C2, respectively; C1 also harbored variants in FRAS1 and NDST2. (B) Schematic representation of the PTCH1 receptor with its extracellular (EC), transmembrane (M), and intra-cellular domains (IC). The positions of the PTCH1 mutations identified in ODA are represented with asterisks, with the color indicating variant pathogenicity. The four variants enclosed with boxes were identified in the initial discovery cohort. p.E2Nfs*9 is specific to isoform NP_001077072 (purple), which is identical to NP_000255 (black) except for an alternate 66 amino acid region at the N terminus.











