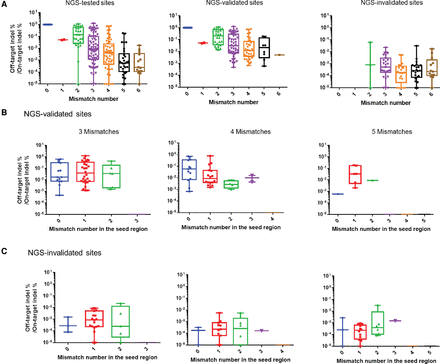
Analysis of NGS-validated and NGS-invalidated off-target sites. Plots of relative indel frequencies (log scale) at off-target sites harboring the number of mismatches indicated in the entire 20-nt sequence (A) or the 10-nt seed sequence (B,C). NGS-tested sites (A) were divided into two groups: validated sites (B) and invalidated sites (C). NGS-validated sites and NGS-invalidated sites were those with indel frequencies above and below, respectively, noise indel levels.











