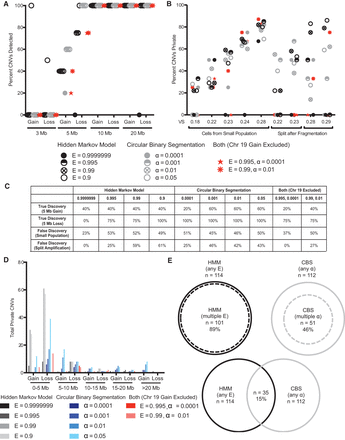
Testing sensitivity and specificity of CNV detection in single-cell sequencing data. (A) The discovery rate of known CNVs ranging from 3 to 20 Mb in single cells using HMM, CBS, or both at different values of E or α. Four or five cells were sequenced for each CNV. (B) The average proportion of CNVs that are private in individual cells sharing a recent ancestor or DNA split after fragmentation from a single cell when analyzed by HMM, CBS, or both at various parameters. (C) The true discovery rates (for 5-Mb gains and losses) and the false discovery rates (averaged across samples from small population or split amplification) for CNV detection using HMM, CBS, or both at different values of E or α, regardless of VS. (D) The distribution of private CNV type and size in individual cells sharing a recent ancestor or DNA split after fragmentation from the same cell when analyzed by HMM, CBS, or both at various parameters. (E) The proportion of private CNVs called by more than one parameter of HMM or CBS and the proportion of private CNVs called by both HMM and CBS.











