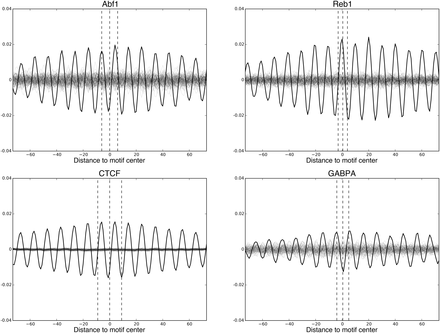
Average oscillations around motif matches of Abf1 and Reb1 in yeast, and CTCF and GABPA in human (“motif oscillations,” black). “Random oscillations” (gray) are average oscillations around randomly chosen genomic sites. One hundred random oscillations are calculated and shown for each TF. Dashed lines indicate the boundaries and centers of the TF motifs. For many TFs, including those shown here, their “motif oscillation” has a significantly higher amplitude than their corresponding “random oscillations,” indicating their TF motif matches locate preferentially with respect to the rotational phasing of the DNA along the nucleosome.











