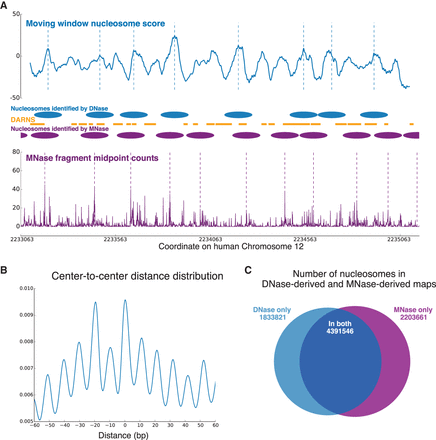
(A) Comparison of DNase-seq–identified nucleosomes and MNase-seq–identified nucleosomes in human Chromosome 12 (intronic region of CACNA1C). Nucleosome centers are identified by the same greedy algorithm on our nucleosome score curve and on the MNase-seq fragment middle point counts, respectively. Nucleosomes identified from the two types of data overlap significantly, especially considering the large noise in the MNase-seq data. The middle panel also shows the DARNS from Winter et al. (2013), which likely represent portions of phased nucleosomes. (B) Distribution of center-to-center distances between our DNase-seq–based nucleosome map and the MNase-seq–based map of Gaffney et al. (2012). (C) The number of nucleosomes shared between the DNase-seq and MNase-seq maps, as well as the number of nucleosomes that appear in only one of the maps.











