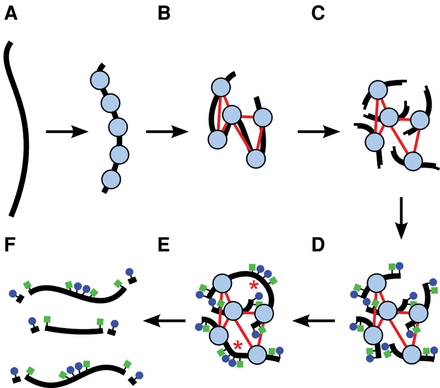
A diagram of a Chicago library generation protocol. (A) Chromatin (nucleosomes in blue) is reconstituted in vitro upon naked DNA (black strand). (B) Chromatin is fixed with formaldehyde (thin red lines are crosslinks). (C) Fixed chromatin is cut with a restriction enzyme, generating free sticky ends (performed on streptavidin-coated beads; data not shown). (D) Sticky ends are filled in with biotinylated (blue circles) and thiolated (green squares) nucleotides. (E) Free blunt ends are ligated (ligations indicated by red asterisks). (F) Crosslinks are reversed and proteins removed to yield library fragments, which are then digested with an exonuclease to remove the terminal biotinylated nucleotides. The thiolated nucleotides protect the interior of the library fragments from digestion.











