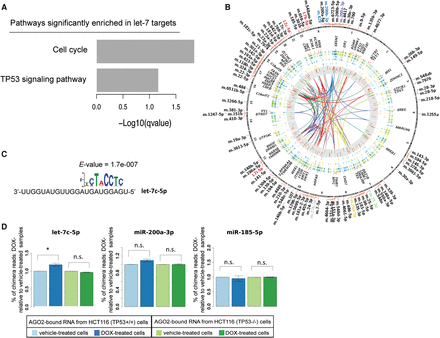
let-7-mRNA chimeras are significantly more abundant after DOX treatment in TP53 wild-type cells compared with untreated cells. (A) KEGG pathways enrichment analysis performed using clusterProfiler (https://www.bioconductor.org/packages/3.3/bioc/html/clusterProfiler.html) (Yu et al. 2012) for the list of genes included in the let-7 subnetwork. (B) Circos (http://circos.ca/) (Krzywinski et al. 2009) circularly layered heatmaps and histograms from different experiment data tracks produced in the present study mapped on the human genome (hg19). From outer to inner circle: heatmap of gene expression changes found in DOX-treated versus vehicle-treated TP53+/+ HCT116 cells by RNA-seq of total and AGO2-bound RNA samples; gene expression changes found in DOX-treated versus vehicle-treated TP53−/− HCT116 cells by RNA-seq of total and AGO2-bound RNA samples; heatmap of miRNA expression changes found in DOX-treated versus vehicle-treated TP53+/+ HCT116 cells by small RNA-seq of total and AGO2-bound RNA samples; miRNA expression changes found in DOX-treated versus vehicle-treated TP53−/− HCT116 cells by small RNA-seq of total and AGO2-bound RNA samples; histogram of changes in PAR-CLIP cluster abundance in DOX-treated versus vehicle-treated samples for HCT116 TP53+/+ cells. Data tracks in the figure render values of observed log-fold changes on a green (lower values) to red (higher values) scale. Ribbons in the circle center show miRNA-target interactions identified by analysis of chimera reads (i.e., ligation products, including part of the mature miRNA and part of its binding site sequence) from the PAR-CLIP experiments. (C) Result of motif analysis performed by using MEME (http://meme.nbcr.net/meme/) (Bailey et al. 2015) on the chimera target sites identified for let-7c. (D) Bar plots of the percentage of total chimera reads in DOX-treated samples compared to vehicle-treated samples.











