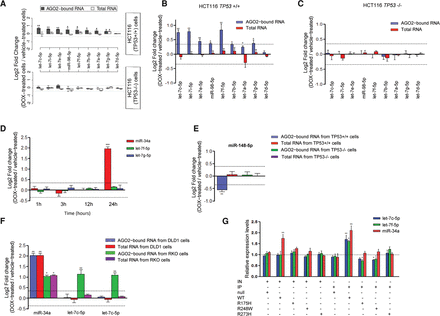
DOX-induced DNA damage induces TP53-dependent differential binding of specific miRNAs to AGO2. (A) Fold changes in let-7 family levels determined by a combined small RNA-seq and AGO2-RIP-seq approach (miRNAs demonstrating a positive or negative fold change equal to or greater than the selected cutoff of 0.35 [log2] and a t-test P-value < 0.05 after correcting for multiple testing with the Benjamini-Hochberg method [Benjamini and Hochberg 1995], were considered to be significant). (B,C) RT-qPCR analysis demonstrates that the binding of let-7 family members onto AGO2 is increased significantly in HCT116 TP53+/+ but not in TP53−/− cells following DOX treatment. (D) Time course showing miR-34a and let-7 family fold changes following DOX-induced DNA damage in total RNA extracted from HCT116 TP53+/+ cells. (E) Fold change in miR-148-5p expression in total RNA or AGO2-bound RNA samples following an AGO2 IP in DOX- or vehicle-treated HCT116 TP53+/+ and TP53−/− cell lines. (F) Fold change in let-7 and miR-34a expression following an AGO2 IP in DLD1 and RKO cell lines following DOX-induced DNA damage. (G) Fold change in let-7 family and miR-34a expression following an AGO2 IP in HCT116 TP53−/− cells transfected with plasmids expressing wild-type or mutant TP53 (R175H, R248W, R273H). (IN) RNA isolated from input samples; (IP) RNA isolated from the immunoprecipitated AGO2; (null) HCT116 TP53−/− cells; (WT) HCT116 TP53−/− cells transfected with the plasmid expressing wild-type TP53; (R175H) HCT116 TP53−/− cells transfected with the plasmid expressing R175H TP53 mutant; (R248W) HCT116 TP53−/− cells transfected with the plasmid expressing R248W TP53 mutant; (R273H) HCT116 TP53−/− cells transfected with the plasmid expressing R273H TP53 mutant. At least three independent experiments have been performed in all cases.











