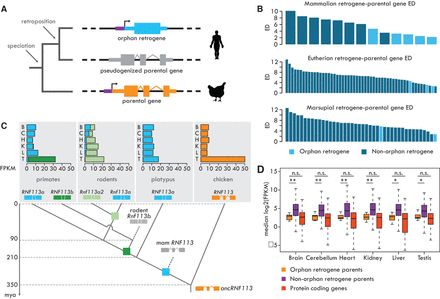
Evolution of orphan retrogenes. (A) Orphan retrogenes are derived from a retroposition event followed by the pseudogenization of the parental gene. (B) Expression divergence between retrogenes and their parental genes, calculated as the Euclidean distance (ED) between the median expression levels across species for each of the six organs. Benjamini-Hochberg-corrected P-values obtained comparing EDs of orphan and other retrogenes with a Mann-Whitney U test. (C, top) Expression profile of the orphan retrogene RNF113 in different clades. For primates and rodents, the expression profiles represent median values across different species. Dark and light green bars indicate the expression levels from two independently originated RNF113 duplicates in primates and rodents, respectively. (B) brain; (C) cerebellum; (H) heart; (K) kidney; (L) liver; (T) testis. (Bottom) Reconstruction of the RNF113 retrogene evolution. Tree nodes indicated by squares correspond to gene duplication events (blue, retroposition; green, duplication mechanism not determined); other nodes correspond to speciation events. (D) Expression profile (median expression across species) of parental genes of orphan and nonorphan retrogenes and annotated protein-coding genes. Significant differences (Mann-Whitney U test with Benjamini-Hochberg correction): (**) P < 0.01; (*) P < 0.05; (n.s.) P > 0.05. Whiskers up to 1.5 times the interquartile range; outliers removed for graphical purposes.











