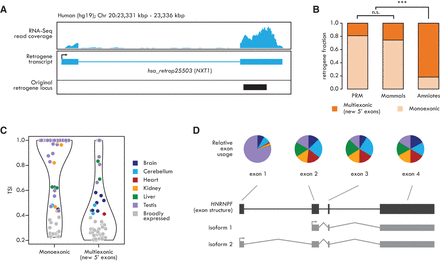
Structural evolution of retrogenes. (A) Transcript structure of the human retrogene hsa_retrop25503 (NXT1) shows the emergence of a new 5′ exon. Black box depicts the original retrocopy locus (coding part). (B) Fractions of human monoexonic and multiexonic (only new 5′ exons) retrogene families from different evolutionary age categories. Significant differences (Fisher's exact test with Benjamini-Hochberg correction): (***) P < 0.001; (n.s.) P > 0.05. (C) Tissue specificity of human monoexonic and multiexonic (only new 5′ exons) retrogenes. The violin plots indicate retrogene TSI distribution; TSI of each retrogene is indicated by colored (when TSI ≥ 0.4, representing tissue with highest expression) or gray (TSI < 0.4) dots. (D, top) Fraction of unique read counts (normalized by the number of reads mapped on the whole gene) from each organ mapping on the human HNRNPF. Exon 1 is significantly more highly transcribed in testis (DEXSeq analysis, Benjamini-Hochberg-corrected P < 0.01). Color code as in C. (Bottom) exon structure (black) and alternative transcripts (gray) of the HNRNPF gene.











