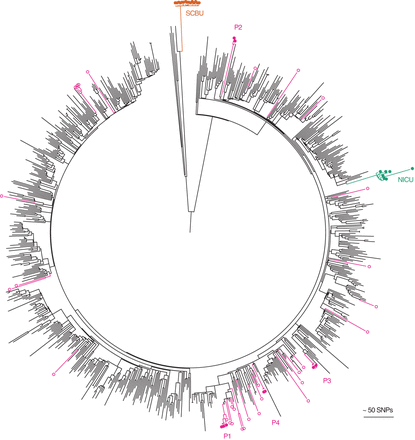
Contextualization of outbreak investigations of CC22 MRSA studied at Cambridge University Hospitals. The maximum likelihood tree was based on 22,238 core SNPs for the 783 ST22 genomes, together with seven isolates from an MRSA outbreak on a neonatal intensive care unit (NICU, green) (Köser et al. 2012), 15 isolates from an MRSA outbreak that focused on a special care baby unit (SCBU, orange) but extended to other wards and the community (Harris et al. 2013), and 42 isolates sequenced as part of an MRSA outbreak investigation on a hepatology ward (nine isolates from four patients with bacteremia [P1-4; pink filled dots] and the remainder from patients who were MRSA carriers on the same ward during a comparable timeframe [pink open dots]) (Török et al. 2014).











