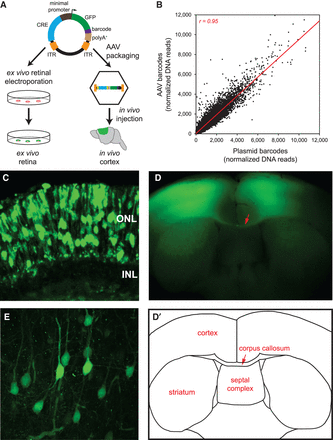
Delivery of capture CRE-seq library into mouse retina ex vivo and cerebral cortex in vivo. (A) Schematic of the CRE-seq library delivery approach. The plasmid library can be directly electroporated into the retina ex vivo. Alternatively, the library can be packaged into AAV and delivered via stereotactic injection into the cerebral cortex in vivo. (B) Scatterplot comparing the relative abundance of approximately 45,000 individual barcoded constructs in the plasmid library delivered into the retina, and in the AAV-packaged library delivered into cortex, as measured by barcode DNA reads summed across the three biological replicates for each tissue and then normalized to the total number of barcode DNA reads. Each data point represents a unique barcoded construct. DNA reads were well-correlated (Pearson r = 0.95), indicating fidelity of barcode representation after AAV packaging and delivery. Off-target constructs and constructs with zero reads in all samples were excluded. Four points falling outside the depicted plot range (included in the calculation of Pearson r) are not shown. (Red line) linear regression. (C) Confocal image of a retina that was electroporated with the plasmid library and cryosectioned after 8 d in culture: (ONL) outer nuclear layer; (INL) inner nuclear layer. (D) Flat-mount image of a coronal slice from a brain injected with the AAV-packaged library bilaterally into the primary motor cortex and harvested ∼4 wk later. (D′) Schematic corresponding to the flat-mount image. Note the bilateral GFP-positive regions in the cortex as well as bundles of GFP-positive axons in the corpus callosum (red arrow). (E) Confocal image of a cortical region infected with the AAV-packaged library.











