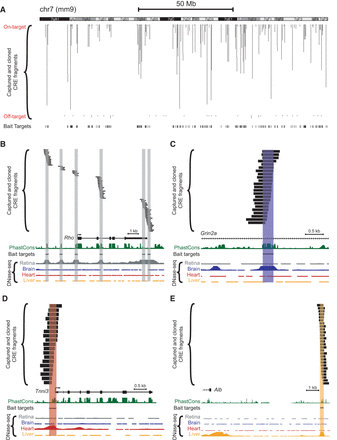
Tiling of captured fragments across target regions. Capture baits were designed based on adult (8-wk-old C57BL/6J) DNase-seq data from Mouse ENCODE (Yue et al. 2014). Paired-end sequencing revealed the locations of individual barcoded, captured-and-cloned fragments. The UCSC Genome Browser (mm9) (Karolchik et al. 2014) screenshots depict: (A) Captured fragments for an entire representative chromosome (Chr 7). Off-target fragments, i.e., those that did not overlap a 300-bp target bait region, are also shown. Examples of captured fragments around a retina-specific locus (B), in an intron of a brain-specific locus (C), in the 5′ UTR/promoter region of a heart-specific locus (D), and downstream from a liver-specific locus (E): (Rho) rhodopsin; (Grin2a) glutamate receptor, ionotropic, NMDA2a (epsilon 1); (Tnni3) troponin I, cardiac 3; (Alb) albumin. Note that some DNase-seq peaks visible in the screenshots were not included as targets for capture. PhastCons depict 30-way vertebrate phylogenetic conservation (Siepel et al. 2005).











