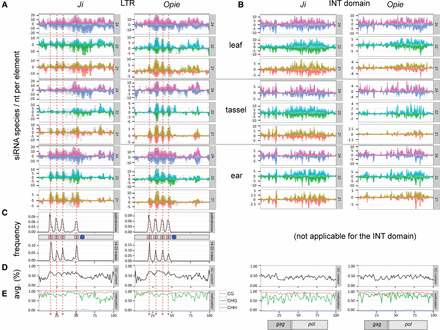
Epigenetic and sequence patterns of Ji and Opie. Each LTR and INT domain was first split in 100 equally sized windows of ∼13-nt and ∼67-nt length, respectively. (A,B) Number of siRNA species per nucleotide along the sense (positive y-axis) and antisense (negative y-axis) strands of the LTRs (A) and INT domain (B) of 3285 and 2926 Ji and Opie elements, respectively (visualized with a box plot for each window). (C) Distribution of palindromes (top) and 19- to 22-nt-long indels (bottom). A schematic shows the approximate positions of the palindromes (pink boxes) and promoter (blue circle). (D) Average GC content. (E) Average methylation level. For the INT domain, the approximate positions of the gag and pol genes are shown.











